JavaBeans example
Shows how to display a set of JavaBeans™ using Rogue Wave® JViews Diagrammer.

Presents an example that displays a chemical molecule.

Describes the classes used to represent atoms, bonds, and molecules.

Describes the class used to connect the
Atom,
Bond, and
Molecule classes to the diagram component.

Describes the class used to represent a data source that contains a Phenol molecule.

Shows how to define a project file that loads the Molecule data source.
The Molecule example
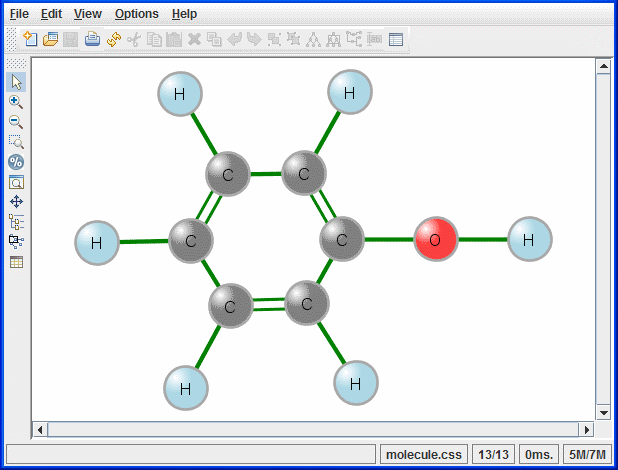
The molecule example displays a chemical molecule. The atoms are the nodes of the graph and the bonds between the atoms are the links of the graph, see the following figure.
The Molecule example: Phenol
The example is supplied with Rogue Wave® JViews Diagrammer in the directory
<installdir>/jviews-diagrammer/codefragments/datamodel/molecule.
The Atom, Bond, and Molecule classes
The atoms and the bonds are represented in the application by the Java ™classes Atom and Bond. These classes obey the JavaBeans™ conventions, for example, the Atom class has a property called symbol which represents the abbreviated symbol of the element; this property can be accessed through the methods setSymbol and getSymbol.
The Atom class
The following code example shows part of the Atom class.
Bean property in the Atom class
/**
* A class that represents an atom.
*/
public class Atom
{
...
private String symbol;
public String getSymbol()
{
return symbol;
}
public void setSymbol(String symbol)
{
this.symbol = symbol;
}
}
The Atom class has also a name property (the name of the element), and an id property, which identifies the atom in the molecule.
The Bond class
The Bond class has two properties firstAtom and secondAtom which contain the identifiers of the two atoms linked by the bond, and also a type property which can have the values single or double.
The Molecule class
A molecule is represented by an instance of the class Molecule. A molecule contains a list of atoms and a list of bonds.
The following code example shows the Molecule class.
Arrays of objects in the Molecule class
public class Molecule
{
private ArrayList atoms = new ArrayList();
private ArrayList bonds = new ArrayList();
public Atom[] getAtoms()
{
return (Atom[]) atoms.toArray(new Atom[0]);
}
public Bond[] getBonds()
{
return (Bond[]) bonds.toArray(new Bond[0]);
}
}
The Molecule model
To connect the existing classes
Atom,
Bond and
Molecule to the diagram component, you need to write a custom SDM model. Since there are already JavaBeans™ that represent the nodes and links of the graph (the
Atom and
Bond classes), the easiest solution is to write a subclass of
IlvJavaBeanSDMModel. This subclass is called
MoleculeModel.
The following code example shows the MoleculeModel class.
public class MoleculeModel extends IlvJavaBeanSDMModel
{
This class represents a molecule, so you can store an instance of the Molecule class as follows:
private Molecule molecule;
The constructor takes the molecule as a parameter. The constructor must also initialize the model to know which bean property holds the node identifier and which bean properties hold the link ends.
The following code example shows the constructor.
public MoleculeModel(Molecule molecule)
{
this.molecule = molecule;
setIDProperty("id");
setFromProperty("firstAtom");
setToProperty("secondAtom");
}
The call to
setIDProperty tells the superclass
IlvJavaBeanSDMModel that it can use the
id property (through the
setId and
getId methods of the
Atom class) to set and retrieve the identifier of a node.
The calls to setFromProperty and setToProperty tell the superclass which properties represent the two end nodes of a link. For example, to retrieve the source node of a link (a Bond instance), the model will call getFirstAtom on the Bond instance.
The model needs to retrieve all the nodes and links of the graph, that is the Atom and Bond objects of the molecule.
The following code example shows the getObjects method, which retrieves the objects.
public Enumeration getObjects()
{
Vector v = new Vector();
Atom[] atoms = molecule.getAtoms();
for (int i = 0; i < atoms.length; i++) {
v.add(atoms[i]);
}
Bond[] bonds = molecule.getBonds();
for (int i = 0; i < bonds.length; i++) {
v.add(bonds[i]);
}
return v.elements();
}
The model must be able to differentiate between nodes and links.
The following code example shows the isLinks method, which returns true for Bond objects and false for Atom objects.
public boolean isLink(Object obj)
{
return obj instanceof Bond;
}
}
The Phenol Molecule data source
The classes and methods of the Molecule example represent a molecule so that the diagram component can display it. To display a specific molecule, you need to load the Molecule instance from a data source.
The following code example shows a data source that contains a Phenol molecule. This class is a subclass of
IlvDiagrammerDataSource.
public class PhenolMoleculeDataSource extends IlvDiagrammerDataSource
{ ...
The Read method
To load a data source into a diagram component, call its read method, which is shown in the following code example.
public void read(IlvDiagrammer diagrammer) throws IlvDiagrammerException
{
Molecule phenol = Molecule.createPhenolMolecule();
MoleculeModel model = new MoleculeModel(phenol);
diagrammer.getEngine().setModel(model);
}
The code lines in the read method are as follows:
1. Create the phenol Molecule instance with a static method for this purpose.
2. Wrap the Molecule instance into the molecule model.
3. Load the new model into the diagram component.
Empty methods
The data source class has other methods which must be implemented because they are abstract, but they do nothing. The following code example shows these methods.
Implementation of unused abstract methods
public void write(IlvDiagrammer diagrammer)
throws IlvDiagrammerException
{
}
protected void serializeImpl(Element element)
throws IlvDiagrammerException
{
}
protected void deserializeImpl(Element element)
throws IlvDiagrammerException
{
}
The write method is not needed because the molecule data cannot be written.
The serialize method is not needed because there is no need to write any additional information to define the data source in a project file.
The deserialize method is not needed because there is no need to read any additional information to load the data source from a project file.
Loading the Molecule into the diagram component
You must define a project file to tell the
diagram component to use the data source. The project file is an XML file called
phenol-molecule.idpr. This file contains the class name of the data source, and also the path of the
style sheet used to represent the molecule.
The following code example shows the project file contents.
The project file for the Molecule example
<diagrammer style="molecule.css">
<datasource class="PhenolMoleculeDataSource"/>
</diagrammer>
To display the molecule, you can simply load the project file into the diagram component as follows:
Diagrammer.setDataFile(new URL("file:phenol-molecule.idpr"));
NOTE The sample uses the prebuilt class IlvDiagrammerApplication, so you do not see the call to setDataFile. Instead the project file is passed as a command-line argument to the application.
Copyright © 2018, Rogue Wave Software, Inc. All Rights Reserved.