LATTICE_DESIGN Function
Analyzes balanced and partially-balanced lattice experiments. In these experiments, a requirement is that the number of treatments be equal to the square of an integer, such as 9, 16, or 25 treatments. Function lattice also analyzes repetitions of lattice experiments.
Usage
result = LATTICE_DESIGN (n, n_locations, n_reps, n_blocks, n_treatments, rep, block, treatment, y)
Input Parameters
n—Number of missing and non-missing experimental observations. LATTICE_DESIGN verifies that:
n = n_locations * t * r
where:
n_locations—Number of locations or repetitions of the lattice experiments. n_locations must be one or greater. If n_locations > 1, then the Locations keyword must be included as input to LATTICE_DESIGN.
n_reps—Number of replicates per location. Each replicate should consist of t = n_treatments organized into 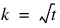 blocks.
blocks.
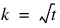 blocks.
blocks. n_blocks—Number of blocks per location. For every location, n_blocks must be equal to n_blocks= r * k, where r = n_reps and 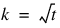 .
.
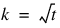 .
.n_treatments—Number of treatments t = n_treatments must be equal to k2.
rep—Array of length n containing the replicate identifiers for each observation in y. For a balanced lattice, the number of replicate identifiers must be equal to n_reps = (k+1). For a partially balanced lattice, the number of replicate identifiers depends upon whether the design is a simple lattice, triple lattice, etc. LATTICE_DESIGN verifies that the number of unique replicate identifiers is equal to n_reps. If multiple locations or repetitions of the experiment are conducted, i.e., n_locations > 1, then the replicate and block numbers contained in rep and block must agree between repetitions.
block—Array of length n containing the block identifiers for each observation in y. LATTICE_DESIGN verifies that the number of unique block identifiers is equal to n_blocks. If multiple locations or repetitions of the experiment are conducted, i.e., n_locations > 1, then block numbers must agree between repetitions. That is, the ith block in every location or repetition must contain the same treatments.
treatment—Array of length n containing the treatment identifiers for each observation in y. Each treatment must be assigned values from 1 to n_treatments. LATTICE_DESIGN verifies that the number of unique treatment identifiers is equal to n_treatments.
y—Array of length n containing the experimental observations and any missing values. Missing values cannot be omitted. They are indicated by placing a NaN (Not a Number) in y. The replicate, block, treatment, and location number for each observation in y are identified by the corrresponding values in the input parameters rep, block, treatment, and the keyword Locations.
Returned Value
result—A two dimensional, 7 by 6 array containing the ANOVA table. Each row in this array contains values for one of the effects in the ANOVA table. The first value in each row, anova_tablei,0 = anova_table(i,0), identifies the source for the effect associated with values in that row. The remaining values in a row contain the ANOVA table values using the convention found in Table 5-39: ANOVA Table Values.
J | anova_tablei,j = anova_table(i,j) |
|---|---|
0 | Source Identifier (values described below) |
1 | Degrees of freedom |
2 | Sum of squares |
3 | Mean squares |
4 | F-statistic |
5 | p-value for this F-statistic |
The Source Identifiers in the first column of anova_tablei,j are the only negative values in anova_table. Assignments of identifiers to ANOVA sources use the coding shown in Table 5-40: Source Identifiers.
Source Identifier | ANOVA Source |
|---|---|
–1 | LOCATIONS* |
–2 | REPLICATES |
–3 | TREATMENTS(unadjusted) |
–4 | TREATMENTS(adjusted) |
–5 | BLOCKS(adjusted) |
–6 | INTRA-BLOCK ERROR |
–7 | CORRECTED TOTAL |
* If N_locations = 1 rows involving location are set to missing (NaN). | |
Input Keywords
Double—If present and nonzero, double precision is used.
Locations—Array of length n containing the location or repetition identifiers for each observation in y. Unique integers must be assigned to each location in the study. This keyword is required when n_locations > 1.
Output Keywords
N_missing—Number of missing values, if any, found in y. Missing values are denoted with a NaN (Not a Number) value.
Cv—The coefficient of variation computed by using the location standard deviation.
Grand_mean—The overall adjusted mean averaged over every location.
Treatment_means—Array of size n_treatments containing the adjusted treatment means.
Std_errors—Array of length 4 containing the standard error and associated degrees of freedom for comparing two treatment means. Std_errors(0) contains the standard error for comparing two treatments that appear in the same block at least once. Std_errors(1) contains the standard error for comparing two treatments that never appear in the same block together. Std_errors(2) contains the standard error for comparing, on average, two treatments from the experiment averaged over cases in which the treatments do or do not appear in the same block. Finally, Std_errors(3) contains the degrees of freedom associated with each of these standard errors, i.e., Std_errors(3)= degrees of freedom for intra-block error.
Location_anova_table—Array of size n_locations by 7 by 6 containing the ANOVA tables associated with each location or repetition of the lattice experiment. For each location, the 7 by 6 dimensional array corresponds to the ANOVA table for that location. For example, Location_anova_table(i,j,k) contains the value in the kth column and jth row of the ANOVA table for the ith location.
Anova_row_labels—Labels for each of the rows of the returned ANOVA table. The label for the ith row of the ANOVA table can be printed with PRINT, Anova_row_labels(i).
Discussion
LATTICE_DESIGN analyzes both balanced and partially-balanced lattice experiments, possibly repeated at multiple locations. These designs were originally described by Yates (1936). A defining characteristic of these classes of lattice experiments is that the number of treatments is always the square of an integer, such as t=9, 16, 25, etc. where t is equal to the number of treatments.
Another characteristic of lattice experiments is that blocks are organized into replicates, where each replicate contains one observation for each treatment. This requires the number of blocks in each replicate to be equal to the number of observations per block. That is, the number of blocks per replicate and the number of observations per block are both equal to 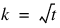 .
.
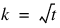 .
.For balanced lattice experiments the number of replicates is always k+1. For partially-balanced lattice experiments, the number of replicates is less than k+1. Tables of balanced-lattice experiments are tabulated in Cochran & Cox (1950) for t=9, 16, 25, 49, 64, and 81.
The analysis of balanced and partially-balanced experiments is detailed in Cochran & Cox (1950) and Kuehl (2000).
Consider, for example, a 3×3 balanced-lattice, i.e., k=3 and t=9. Notice that the number of replicates is 4 and the number of blocks per replicate is equal to 3. The total number of blocks is equal to:
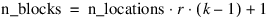
For a balanced-lattice:
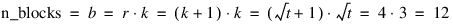
Replicate I | Replicate II |
|---|---|
Block 1 (T1, T2, T3) | Block 4 (T1, T4, T7) |
Block 2 (T4, T5, T6) | Block 5 (T2, T5, T8) |
Block 3 (T7, T8, T9) | Block 6 (T3, T6, T9) |
Replicate III | Replicate IV |
Block 7 (T1, T5, T9) | Block 10 (T1, T6, T8) |
Block 8 (T2, T6, T7) | Block 11 (T2, T4, T9) |
Block 9 (T3, T4, T8) | Block 12 (T3, T5, T7) |
The analysis of variance for data from a balanced-lattice experiment, takes the form familiar to other balanced incomplete block experiments. In these experiments, the error term is divided into two components: the Inter-Block Error and the Intra-Block Error. For single and multiple locations, the general format of the ANOVA tables is illustrated in the Table 5-42: ANOVA Table for a Lattice Experiment at one Location and Table 5-43: ANOVA Table for a Lattice Experiment at Multiple Locations.
SOURCE | DF | Sum of Squares | Mean Squares |
|---|---|---|---|
REPLICATES | r – 1 | SSR | MSR |
TREATMENTS(unadj) | t – 1 | SST | MST |
TREATMENTS(adj) | t – 1 | SSTa | MSTa |
BLOCKS(adj) | 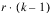 | SSBa | MSBa |
INTRA-BLOCK ERROR | 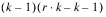 | SSI | MSI |
TOTAL | 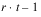 | SSTot | |
SOURCE | DF | Sum of Squares | Mean Squares |
|---|---|---|---|
LOCATIONS | p – 1 | SSL | MSL |
REPLICATES WITHIN LOCATIONS | p(r – 1) | SSR | MSR |
TREATMENTS(unadj) | t – 1 | SST | MST |
TREATMENTS(adj) | t – 1 | SSTa | MSTa |
BLOCKS(adj) | 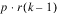 | SSB | MSB |
INTRA-BLOCK ERROR | 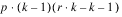 | SSI | MSI |
TOTAL | 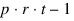 | SSTot | |
Example 1
This example is a lattice design for 16 treatments conducted at one location. A lattice design with t = k2 = 16 treatments is a balanced lattice design with r = k+1 = 5 replicates and r·k = 5(4) = 20 blocks.
; Total number of observations
n = 80
; Number of locations
n_locations = 1
; Number of treatments
n_treatments = 16
; Number of replicates
n_reps = 5
; Total number of blocks
n_blocks = 20
rep = [ $
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, $
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, $
3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, $
4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, $
5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5 ]
block = [ $
1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, $
4, 4, 4, 4, 5, 5, 5, 5, 6, 6, 6, 6, $
7, 7, 7, 7, 8, 8, 8, 8, 9, 9, 9, 9, $
10, 10, 10, 10, 11, 11, 11, 11, 12, 12, 12, 12, $
13, 13, 13, 13, 14, 14, 14, 14, 15, 15, 15, 15, $
16, 16, 16, 16, 17, 17, 17, 17, 18, 18, 18, 18, $
19, 19, 19, 19, 20, 20, 20, 20 ]
treatments = [ $
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, $
14, 15, 16, 1, 5, 9, 13, 10, 2, 14, 6, 7, 15, $
3, 11, 16, 8, 12, 4, 1, 6, 11, 16, 5, 2, 15, $
12, 9, 14, 3, 8, 13, 10, 7, 4, 1, 14, 7, 12, $
13, 2, 11, 8, 5, 10, 3, 16, 9, 6, 15, 4, 1, $
10, 15, 8, 9, 2, 7, 16, 13, 6, 3, 12, 5, 14, $
11, 4 ]
y = [ $
147, 152, 167, 150, 127, 155, 162, 172, $
147, 100, 192, 177, 155, 195, 192, 205, $
140, 165, 182, 152, 97, 155, 192, 142, $
155, 182, 192, 192, 182, 207, 232, 162, $
155, 132, 177, 152, 182, 130, 177, 165, $
137, 185, 152, 152, 185, 122, 182, 192, $
220, 202, 175, 205, 205, 152, 180, 187, $
165, 150, 200, 160, 155, 177, 185, 172, $
147, 112, 177, 147, 180, 205, 190, 167, $
172, 212, 197, 192, 177, 220, 205, 225 ]
aov = LATTICE_DESIGN(n, n_locations, n_reps, n_blocks, $
n_treatments, rep, block, treatments, $
y, Grand_mean=grand_mean, Cv=cv, $
Treatment_means=treatment_means, $
Std_err=std_err)
labels = ['Locations ', $
'Replicates ', $
'Treatments ', $
' (unadjusted)' , $
'Treatments ', $
' (adjusted)' , $
'Blocks (adjusted)', $
'Intra-Block Error', $
'Corrected Total ']
; Print Analysis of Variance Table
PRINT, " *** ANALYSIS OF VARIANCE TABLE ***"
PRINT, 'ID', 'DF', 'SSQ', 'MS', 'F-test', 'P-Value', $
Format='(A21, A7, A8, A9, A8, A8)' & $
idx = 0
FOR i=0L, (SIZE(aov))(1)-1 DO BEGIN & $
PRINT, labels(idx), aov(i,0), aov(i,1), $
aov(i,2), aov(i,3), aov(i,4), aov(i,5), Format= $
'(A17, 1X, I3, 2X, F3.0, 2X, F8.2, 2X, F7.2, 2X, ' + $
'F5.2, 2X, F7.3)' & $
idx = idx + 1 & $
IF idx LT N_ELEMENTS(labels)-1 THEN $
WHILE STRPOS(labels(idx), ' ', 0) EQ 0 DO BEGIN & $
PRINT, labels(idx) & idx = idx + 1 & $
ENDWHILE & $
ENDFOR
PRINT, ''
PRINT, grand_mean, $
Format='("Adjusted Grand Mean :", F8.3)'PRINT, cv, Format='("Coefficient of Variation:", F8.3)'PRINT, ''
PRINT, "Adjusted Treatment Means:"
FOR i=0L, n_treatments-1 DO $
PRINT, (i+1), treatment_means(i), Format= $
"(2X, 'Treatment[', I2, '] Mean:', F10.4)"
PRINT, ''
PRINT, std_err(0), $
Format='("Standard Error for Comparing Two Treatment ' + $'Means: ", F9.6, I1)'
PRINT, FIX(std_err(3)), Format='("(df=", I2, ")")'PRINT, ''
; Perform multiple comparison using the LSD procedure
equal_means = MULTICOMP(treatment_means, $
std_err(3), std_err(2)/SQRT(2.0), $
/LSD, Alpha=0.05)
PM, equal_means, $
Title="LSD Comparison: Size of Groups of Means"
Output
*** ANALYSIS OF VARIANCE TABLE ***
ID DF SSQ MS F-test P-Value
Locations -1 NaN NaN NaN NaN NaN
Replicates -2 4. 6524.50 1631.12 NaN NaN
Treatments -3 15. 27297.00 1819.80 4.12 0.000
(unadjusted)
Treatments -4 15. 21271.20 1418.08 4.21 0.000
(adjusted)
Blocks (adjusted) -5 15. 11339.28 755.95 NaN NaN
Intra-Block Error -6 45. 15173.23 337.18 NaN NaN
Corrected Total -7 79. 60334.00 NaN NaN NaN
Adjusted Grand Mean : 171.450
Coefficient of Variation: 10.710
Adjusted Treatment Means:
Treatment[ 1] Mean: 166.4533
Treatment[ 2] Mean: 160.7527
Treatment[ 3] Mean: 183.6289
Treatment[ 4] Mean: 175.6298
Treatment[ 5] Mean: 162.6807
Treatment[ 6] Mean: 167.6716
Treatment[ 7] Mean: 168.3822
Treatment[ 8] Mean: 176.5731
Treatment[ 9] Mean: 162.6928
Treatment[10] Mean: 118.5197
Treatment[11] Mean: 189.0615
Treatment[12] Mean: 190.4608
Treatment[13] Mean: 169.4514
Treatment[14] Mean: 197.0827
Treatment[15] Mean: 185.3560
Treatment[16] Mean: 168.8029
Standard Error for Comparing Two Treatment Means: 13.221848
(df=45)
LSD Comparison: Size of Groups of Means
0
12
12
0
11
0
0
0
0
7
0
0
0
0
0
Example 2
This example consists of a 5 × 5 partially-balanced lattice repeated twice. In this case, the number of replicates is not k+1 = 6, it is only n_reps = 2. Each lattice consists of total of 50 observations which is repeated twice. The first observation in this experiment is missing.
a = MACHINE(/Float)
NaN = a.NAN
; Total number of observations
n = 100
; Number of locations
n_locations = 2
; Number of treatments
n_treatments = 25
; Number of replicates
n_reps = 2
; Total number of blocks
n_blocks = 10
rep = [ $
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, $
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, $
1, 1, 1, 1, 1, 2, 2, 2, 2, 2, $
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, $
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, $
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, $
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, $
1, 1, 1, 1, 1, 2, 2, 2, 2, 2, $
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, $
2, 2, 2, 2, 2, 2, 2, 2, 2, 2 ]
block = [ $
1, 1, 1, 1, 1, 2, 2, 2, 2, 2, $
3, 3, 3, 3, 3, 4, 4, 4, 4, 4, $
5, 5, 5, 5, 5, 6, 6, 6, 6, 6, $
7, 7, 7, 7, 7, 8, 8, 8, 8, 8, $
9, 9, 9, 9, 9, 10, 10, 10, 10, 10, $
1, 1, 1, 1, 1, 2, 2, 2, 2, 2, $
3, 3, 3, 3, 3, 4, 4, 4, 4, 4, $
5, 5, 5, 5, 5, 6, 6, 6, 6, 6, $
7, 7, 7, 7, 7, 8, 8, 8, 8, 8, $
9, 9, 9, 9, 9, 10, 10, 10, 10, 10 ]
treatment = [ $
1, 2, 3, 4, 5, $
6, 7, 8, 9, 10, $
11, 12, 13, 14, 15, $
16, 17, 18, 19, 20, $
21, 22, 23, 24, 25, $
1, 6, 11, 16, 21, $
2, 7, 12, 17, 22, $
3, 8, 13, 18, 23, $
4, 9, 14, 19, 24, $
5, 10, 15, 20, 25, $
1, 2, 3, 4, 5, $
6, 7, 8, 9, 10, $
11, 12, 13, 14, 15, $
16, 17, 18, 19, 20, $
21, 22, 23, 24, 25, $
1, 6, 11, 16, 21, $
2, 7, 12, 17, 22, $
3, 8, 13, 18, 23, $
4, 9, 14, 19, 24, $
5, 10, 15, 20, 25 ]
locations = [ $
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, $
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, $
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, $
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, $
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, $
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, $
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, $
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, $
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, $
2, 2, 2, 2, 2, 2, 2, 2, 2, 2 ]
y = [$
NaN, 7, 5, 8, 6, $
16, 12, 12, 13, 8, $
17, 7, 7, 9, 14, $
18, 16, 13, 13, 14, $
14, 15, 11, 14, 14, $
24, 13, 24, 11, 8, $
21, 11, 14, 11, 23, $
16, 4, 12, 12, 12, $
17, 10, 30, 9, 23, $
15, 15, 22, 16, 19, $
13, 26, 9, 13, 11, $
15, 18, 22, 11, 15, $
19, 10, 10, 10, 16, $
21, 16, 17, 4, 17, $
15, 12, 13, 20, 8, $
16, 7, 20, 13, 21, $
15, 10, 11, 7, 14, $
7, 11, 15, 15, 16, $
19, 14, 20, 6, 16, $
17, 18, 20, 15, 14 ]
aov = LATTICE_DESIGN(n, n_locations, n_reps, n_blocks, $
n_treatments, rep, block, treatment, $
y, Locations=locations, $
Cv=cv, $
Grand_mean=grand_mean, $
Location_anova_table= $
location_anova_table, $
Treatment_means=treatment_means, $
Anova_row_labels=anova_row_labels, $
Std_err=std_err, $
N_missing=n_missing)
labels = ['Locations ', $
'Replicates within', $
' Locations' , $
'Treatments ', $
' (unadjusted)' , $
'Treatments ', $
' (adjusted)' , $
'Blocks (adjusted)', $
'Intra-Block Error', $
'Corrected Total ']
; Print Analysis of Variance Table
PRINT, " *** ANALYSIS OF VARIANCE TABLE ***"
PRINT, 'ID', 'DF', 'SSQ', 'MS', 'F-test', 'P-Value', $
Format='(A21, A7, A8, A9, A8, A8)' & $
idx = 0
FOR i=0L, (SIZE(aov))(1)-1 DO BEGIN & $
PRINT, labels(idx), aov(i,0), aov(i,1), $
aov(i,2), aov(i,3), aov(i,4), aov(i,5), Format= $
'(A17, 1X, I3, 2X, F3.0, 2X, F8.2, 2X, F7.2, 2X, ' + $
'F5.2, 2X, F7.3)' & $
idx = idx + 1 & $
IF idx LT N_ELEMENTS(labels)-1 THEN $
WHILE STRPOS(labels(idx), ' ', 0) EQ 0 DO BEGIN & $
PRINT, labels(idx) & idx = idx + 1 & $
ENDWHILE & $
ENDFOR
PRINT, ''
; Print the location ANOVA tables
idx = 0
FOR j=0L, n_locations-1 DO BEGIN & $
PRINT, "LOCATION", j, Format='(A33, 1X, I1)' & $
PRINT, "*** ANALYSIS OF VARIANCE TABLE ***", $
Format='(A47)' & $
PRINT, 'ID', 'DF', 'SSQ', 'MS', 'F-test', 'P-Value', $
Format='(A23, A5, A9, A8, A8, A8)' & $
FOR i=0L, (SIZE(aov))(1)-1 DO BEGIN & $
PRINT, labels(idx), location_anova_table(j,i,0), $
location_anova_table(j,i,1), $
location_anova_table(j,i,2), $
location_anova_table(j,i,3), $
location_anova_table(j,i,4), $
location_anova_table(j,i,5), Format= $
'(A17, 2X, I3, 3X, F3.0, 2X, F7.2, 2X, F6.2, ' + $
'2X, F5.2, 2X, F7.3)' & $
idx = idx + 1 & $
IF idx LT N_ELEMENTS(labels)-1 THEN $
WHILE STRPOS(labels(idx), ' ', 0) EQ 0 DO BEGIN & $
PRINT, labels(idx) & idx = idx + 1 & $
ENDWHILE & $
ENDFOR & $
idx = 0 & $
PRINT, '' & $
ENDFOR
PRINT, ''
PRINT, grand_mean, $
Format='("Adjusted Grand Mean :", F8.3)'PRINT, cv, Format='("Coefficient of Variation:", F8.3)'PRINT, ''
PRINT, "Adjusted Treatment Means:"
FOR i=0L, n_treatments-1 DO $
PRINT, (i+1), treatment_means(i), Format= $
"(2X, 'Treatment[', I2, '] Mean:', F10.4)"
PRINT, ''
PRINT, std_err(2), $
Format='("Standard Error for Comparing Two Treatment ' + $'Means: ", F9.6, I1)'
PRINT, FIX(std_err(3)), Format='("(df=", I2, ")")'PRINT, ''
; Perform multiple comparison using the LSD procedure
equal_means = MULTICOMP(treatment_means, std_err(3), $
std_err(2)/SQRT(2.0), $
/LSD, Alpha=0.05)
PM, equal_means, $
Title="LSD Comparison: Size of Groups of Means"
PRINT, ''
PRINT, n_missing, Format= $
'("Number of missing observations: ", I2)'Output
*** ANALYSIS OF VARIANCE TABLE ***
ID DF SSQ MS F-test P-Value
Locations -1 1. 12.19 12.19 0.25 0.622
Replicates within -2 2. 203.99 101.99 7.44 0.001
Locations
Treatments -3 24. 795.46 33.14 0.02 1.000
(unadjusted)
Treatments -4 24. 951.20 39.63 2.89 0.006
(adjusted)
Blocks (adjusted) -5 16. 770.50 48.16 3.51 0.000
Intra-Block Error -6 55. 753.82 13.71 NaN NaN
Corrected Total -7 98. 2535.95 NaN NaN NaN
LOCATION 0
*** ANALYSIS OF VARIANCE TABLE ***
ID DF SSQ MS F-test P-Value
Locations -1 NaN NaN NaN NaN NaN
Replicates within -2 1. 203.67 203.67 NaN NaN
Locations
Treatments -3 24. 567.13 23.63 0.78 0.721
(unadjusted)
Treatments -4 24. 661.08 27.54 2.04 0.078
(adjusted)
Blocks (adjusted) -5 8. 490.51 61.31 NaN NaN
Intra-Block Error -6 15. 202.93 13.53 NaN NaN
Corrected Total -7 48. 1464.24 NaN NaN NaN
LOCATION 1
*** ANALYSIS OF VARIANCE TABLE ***
ID DF SSQ MS F-test P-Value
Locations -1 NaN NaN NaN NaN NaN
Replicates within -2 1. 0.32 0.32 NaN NaN
Locations
Treatments -3 24. 622.52 25.94 1.43 0.196
(unadjusted)
Treatments -4 24. 707.51 29.48 2.83 0.018
(adjusted)
Blocks (adjusted) -5 8. 269.76 33.72 NaN NaN
Intra-Block Error -6 16. 166.92 10.43 NaN NaN
Corrected Total -7 49. 1059.52 NaN NaN NaN
Adjusted Grand Mean : 14.011
Coefficient of Variation: 26.423
Adjusted Treatment Means:
Treatment[ 1] Mean: 17.1507
Treatment[ 2] Mean: 19.2200
Treatment[ 3] Mean: 11.1261
Treatment[ 4] Mean: 14.6230
Treatment[ 5] Mean: 12.6543
Treatment[ 6] Mean: 11.8133
Treatment[ 7] Mean: 11.9045
Treatment[ 8] Mean: 11.3106
Treatment[ 9] Mean: 9.5576
Treatment[10] Mean: 11.5889
Treatment[11] Mean: 22.1320
Treatment[12] Mean: 12.7232
Treatment[13] Mean: 13.1293
Treatment[14] Mean: 17.8763
Treatment[15] Mean: 18.6576
Treatment[16] Mean: 14.6568
Treatment[17] Mean: 11.4980
Treatment[18] Mean: 13.1540
Treatment[19] Mean: 5.4010
Treatment[20] Mean: 12.9323
Treatment[21] Mean: 15.4108
Treatment[22] Mean: 17.0020
Treatment[23] Mean: 13.9081
Treatment[24] Mean: 17.6550
Treatment[25] Mean: 13.1864
Standard Error for Comparing Two Treatment Means: 4.617282
(df=55)
LSD Comparison: Size of Groups of Means
16
22
22
0
0
0
0
0
0
0
15
0
0
0
0
0
0
0
0
0
0
0
0
0
Number of missing observations: 1