LATIN_SQUARE Function
Analyzes data from latin-square experiments. LATIN_SQUARE also analyzes latin-square experiments replicated at several locations.
Usage
result = latin_square (n, n_locations, n_treatments, row, col, treatment, y)
Input Parameters
n—Number of missing and non-missing experimental observations. LATIN_SQUARE verifies that:
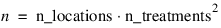
n_locations—Number of locations. Must be greater than or equal to one. If n_locations > 1 then the Locations keyword must be included as input to LATIN_SQUARE.
n_treatments—Number of treatments. n_treatments must be greater than one. In addition, the number of unique row and column identifiers specified in the input parameters row and col must be equal to n_treatments.
row—Array of length n containing the row identifiers for each observation in y. Each row must be assigned values from 1 to n_treatments. LATIN_SQUARE verifies that the number of unique factor A identifiers is equal to n_treatments.
col—Array of length n containing the column identifiers for each observation in y. Each column must be assigned values from 1 to n_treatments. LATIN_SQUARE verifies that the number of unique column identifiers is equal to n_treatments.
treatment—Array of length n containing the treatment identifiers for each observation in y. Each treatment must be assigned values from 1 to n_treatments. LATIN_SQUARE verifies that the number of unique treatment identifiers is equal to n_treatments.
y—Array of length n containing the experimental observations and any missing values. Missing values cannot be omitted. They are indicated by placing a NaN (Not a Number) at the appropriate positions in y. NaN can be defined by calling the MACHINE function. For example:
x = MACHINE(/Float)
y(i) = x.NaN
The location, row, column, and treatment number for each observation in y are identified by the corresponding values in Locations, row, col, and n_treatments.
Returned Value
result—A two dimensional, 7 by 6 array containing the ANOVA table. Each row in this array contains values for one of the effects in the ANOVA table. The first value in each row, anova_tablei,0 = anova_table(i,0), identifies the source for the effect associated with values in that row. The remaining values in a row contain the ANOVA table values shown in Table 5-34: ANOVA Table Values.
J | anova_tablei,j = anova_table(i,j) |
|---|---|
0 | Source Identifier (values described below) |
1 | Degrees of freedom |
2 | Sum of squares |
3 | Mean squares |
4 | F-statistic |
5 | p-value for this F-statistic |
The Source Identifiers in the first column of anova_tablei,j are the only negative values in anova_table. Assignments of identifiers to ANOVA sources use the coding shown in Table 5-35: ANOVA Assignment Identifiers:
Source Identifier | ANOVA Source |
|---|---|
–1 | LOCATIONS* |
–2 | ROWS |
–3 | COLUMNS |
–4 | TREATMENTS |
–5 | LOCATIONS × TREATMENTS* |
–6 | ERROR WITHIN LOCATIONS |
–7 | CORRECTED TOTAL |
* If n_locations = 1 rows involving location are set to missing (NaN). | |
Input Keywords
Double—If present and nonzero, double precision is used.
Locations—Array of length n containing the location identifiers for each observation in y. Unique integers must be assigned to each location in the study. This keyword is required when n_locations > 1.
Output Keywords
N_missing—Number of missing values, if any, found in y. Missing values are denoted with a NaN (Not a Number) value.
Cv—The coefficient of variation computed by using the within location standard deviation.
Grand_mean—Mean of all the data across every location.
Treatment_means—Array of size n_treatments containing the treatment means.
Std_errors—Array of length two containing the standard error and associated degrees of freedom for comparing two treatment means. Std_errors(0) contains the standard error and its degrees of freedom are returned in Std_errors(1).
Location_anova_table—Array of size n_locations by 7 by 6 containing the ANOVA tables associated with each location. For each location, the 7 by 6 dimensional array corresponds to the ANOVA table for that location. For example, Location_anova_table(i, j, k) contains the value in the kth column and jth row of the ANOVA table for the ith location.
Anova_row_labels—Labels for each of the 7 rows of the returned ANOVA table. The label for the ith row of the ANOVA table can be printed with PRINT, Anova_row_labels(i).
Discussion
LATIN_SQUARE analyzes latin-square experiments, possibly replicated at multiple locations. Latin-square experiments block treatments using two factors: rows and columns. The number of levels associated with rows and columns must equal the number of treatments. Treatments are blocked by rows and columns in a balanced arrangement to ensure that every row contain one replicate of every treatment. The same balance is required for every column, see Table 5-36: Latin-Square Experiment with Four Treatments. Notice that the four treatments, T1, T2, T3, and T4, appear exactly once in every column and every row.
Columns | |||||
C1 | C2 | C3 | C4 | ||
Rows | R1 | T1 | T2 | T3 | T4 |
R2 | T2 | T3 | T4 | T1 | |
R3 | T3 | T4 | T1 | T2 | |
R4 | T4 | T1 | T2 | T3 | |
A necessary assumption in Latin-Square experiments is that there are no interactions between treatments and the row and column blocking factors. For data collected at a single location, the ANOVA table for a Latin-Square experiment is usually organized into five rows, see Table 5-37: ANOVA Table for a Latin-Square Experiment at one Location.
SOURCE | DF | Sum of Squares | Mean Squares |
|---|---|---|---|
ROWS | t – 1 | 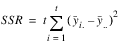 | MSR |
COLUMNS | t – 1 | 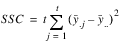 | MSC |
TREATMENTS | t – 1 | 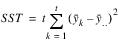 | MST |
ERROR | (t – 1)(t – 2) | SSE=SSTot-SSR-SSC-SST | MSE |
TOTAL | t2 – 1 | 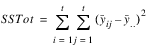 | |
The statistical model used to represent data is from a single location:
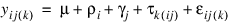
where:
If multiple locations are involved, LATIN_SQUARE assumes that treatments are crossed with locations, but that row and column effects are nested within locations, see Table 5-38: ANOVA Table for a Latin-Square Experiment at Multiple Locations. The statistical model used to represent this data is:
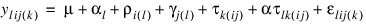
where:
SOURCE | DF | Sum of Squares | Mean Squares |
|---|---|---|---|
LOCATIONS | r – 1 | 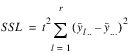 | MSL |
ROWS | r (t – 1) | 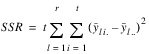 | MSR |
COLUMNS | r (t – 1) | 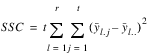 | MSC |
TREATMENTS | t – 1 | 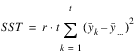 | MST |
LOCATIONS X TREATMENTS | (r – 1)(t – 1) | SSLT by difference | MSLT |
ERROR | (t – 1)(r(t – 1) – 1) | 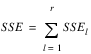 | MSE |
TOTAL | 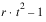 | 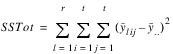 |
Example
This example uses four treatments organized into a latin square.
; Total number of observations
n = 16
; Number of locations
n_locations = 1
; Number of rows, columns and treatments
n_treatments = 4
col = [1, 2, 3, 4, 1, 2, 3, 4, 1, 2, 3, 4, 1, 2, 3, 4]
row = [3, 2, 4, 1, 1, 4, 2, 3, 2, 3, 1, 4, 4, 1, 3, 2]
treatment = [1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4]
y = [1.167, 1.185, 1.655, 1.345, 1.64 , 1.29, 1.665, 1.29, $
1.475, 0.71, 1.425, 0.66, 1.565, 1.29, 1.4 , 1.18]
aov = LATIN_SQUARE(n, n_locations, n_treatments, row, col, $
treatment, y, cv=cv, $
treatment_means=treatment_means, $
std_err=std_err, $
grand_mean=grand_mean, $
anova_row_labels=anova_row_labels)
PRINT, "*** Experimental Design ***"
PRINT, "========================"
PRINT, "|COL | 1 | 2 | 3 | 4 |"
PRINT, "========================"
PRINT, "|ROW 1 | 2 | 4 | 3 | 1 |"
PRINT, "========================"
PRINT, "|ROW 2 | 3 | 1 | 2 | 4 |"
PRINT, "========================"
PRINT, "|ROW 3 | 1 | 3 | 4 | 2 |"
PRINT, "========================"
PRINT, "|ROW 4 | 4 | 2 | 1 | 3 |"
PRINT, "========================"
PRINT, ''
; Print Analysis of Variance Table
PRINT, " *** ANALYSIS OF VARIANCE TABLE ***"
PRINT, 'ID', 'DF', 'SSQ', 'MS', 'F-test', 'P-Value', $
Format='(A27, A5, A7, A6, A8, A8)' & $
FOR i=0L, (SIZE(aov))(1)-1 DO $
PRINT, anova_row_labels(i), aov(i,0), aov(i,1), $
aov(i,2), aov(i,3), aov(i,4), aov(i,5), Format= $
'(A23, 1X, I3, 2X, F3.0, 1X, F6.2, 1X, F6.2, 2X, ' + $
'F5.2, 2X, F6.3)'
PRINT, ''
PRINT, grand_mean, $
Format='("Grand Mean :", F7.3)'PRINT, cv, Format='("Coefficient of Variation:", F7.3)'PRINT, ''
PRINT, "Treatment Means:"
FOR i=0L, n_treatments-1 DO $
PRINT, (i+1), treatment_means(i), Format= $
"(2X, 'Treatment[', I1, '] Mean:', F7.4)"
PRINT, ''
PRINT, std_err(0), Format='("Standard Error for ' + $'Comparing Two Treatment Means: ", F8.6, I1)'
PRINT, FIX(std_err(1)), Format='("(df=", I1, ")")'PRINT, ''
; Perform multiple comparison using the LSD procedure
equal_means = MULTICOMP(treatment_means, std_err(1), $
std_err(0)/SQRT(2.0), $
/LSD, Alpha=0.05)
PM, equal_means, $
Title="LSD Comparison: Size of Groups of Means"
Output
*** Experimental Design ***
========================
|COL | 1 | 2 | 3 | 4 |
========================
|ROW 1 | 2 | 4 | 3 | 1 |
========================
|ROW 2 | 3 | 1 | 2 | 4 |
========================
|ROW 3 | 1 | 3 | 4 | 2 |
========================
|ROW 4 | 4 | 2 | 1 | 3 |
========================
*** ANALYSIS OF VARIANCE TABLE ***
ID DF SSQ MS F-test P-Value
Locations ............. -1 NaN NaN NaN NaN NaN
Rows within Locations . -2 3. 0.18 0.06 2.06 0.207
Columns within Location -3 3. 0.59 0.20 6.58 0.025
Treatments ............ -4 3. 0.35 0.12 3.93 0.073
Locations x Treatments -5 NaN NaN NaN NaN NaN
Error within Locations -6 6. 0.18 0.03 NaN NaN
Corrected Total ....... -7 15. 1.31 NaN NaN NaN
Grand Mean : 1.309
Coefficient of Variation: 13.204
Treatment Means:
Treatment[1] Mean: 1.3380
Treatment[2] Mean: 1.4712
Treatment[3] Mean: 1.0675
Treatment[4] Mean: 1.3587
Standard Error for Comparing Two Treatment Means: 0.122202
(df=6)
LSD Comparison: Size of Groups of Means
3
3
0